Using tidy data with Bayesian models
Matthew Kay
2024-09-14
Source:vignettes/tidybayes.Rmd
tidybayes.RmdIntroduction
This vignette introduces the tidybayes package, which
facilitates the use of tidy data (one observation per row) with Bayesian
models in R. This vignette is geared towards working with tidy data in
general-purpose modeling functions like JAGS or Stan. For a similar
introduction to the use of tidybayes with high-level
modeling functions such as those in brms or
rstanarm, see vignette("tidy-brms") or
vignette("tidy-rstanarm"). This vignette also describes how
to use ggdist (the sister package to
tidybayes) for visualizing model output.
The default output (and sometimes input) data formats of popular
modeling functions like JAGS and Stan often don’t quite conform to the
ideal of tidy
data. For example, input formats might expect a list instead of a
data frame, and for all variables to be encoded as numeric values
(requiring translation of factors to numeric values and the creation of
index variables to store the number of levels per factor or the number
of observations in a data frame). Output formats will often be in matrix
form (requiring conversion for use with libraries like ggplot), and will
use numeric indices (requiring conversion back into factor level names
if the you wish to make meaningfully-labeled plots or tables).
tidybayes automates all of these sorts of tasks.
Philosophy
There are a few core ideas that run through the
tidybayes API that should (hopefully) make it easy to
use:
Tidy data does not always mean all parameter names as values. In contrast to the
ggmcmclibrary (which translates model results into a data frame with aParameterandvaluecolumn), thespread_drawsfunction intidybayesproduces data frames where the columns are named after parameters and (in some cases) indices of those parameters, as automatically as possible and using a syntax as close to the same way you would refer to those variables in the model’s language as possible. A similar function toggmcmc’s approach is also provided ingather_draws, since sometimes you do want variable names as values in a column. The goal is fortidybayesto do the tedious work of figuring out how to make a data frame look the way you need it to, including turning parameters with indices like"b[1,2]"and the like into tidy data for you.Fit into the tidyverse.
tidybayesmethods fit into a workflow familiar to users of thetidyverse(dplyr,tidyr,ggplot2, etc), which means fitting into the pipe (%>%) workflow, using and respecting grouped data frames (thusspread_drawsandgather_drawsreturn results already grouped by variable indices, and methods likemedian_qicalculate point summaries and intervals for variables and groups simultaneously), and not reinventing too much of the wheel if it is already made easy by functions provided by existingtidyversepackages (unless it makes for much clearer code for a common idiom). For compatibility with other package column names (such asbroom::tidy),tidybayesprovides transformation functions liketo_broom_namesthat can be dropped directly into data transformation pipelines.Focus on composable operations and plotting primitives, not monolithic plots and operations. Several other packages (notably
bayesplotandggmcmc) already provide an excellent variety of pre-made methods for plotting Bayesian results.tidybayesshies away from duplicating this functionality. Instead, it focuses on providing composable operations for generating and manipulating Bayesian samples in a tidy data format, and graphical primitives forggplotthat allow you to build custom plots easily. Most simply, wherebayesplotandggmcmctend to have functions with many options that return a full ggplot object,tidybayestends towards providing primitives (likegeoms) that you can compose and combine into your own custom plots. I believe both approaches have their place: pre-made functions are especially useful for common, quick operations that don’t need customization (like many diagnostic plots), while composable operations tend to be useful for more complex custom plots (in my opinion).Sensible defaults make life easy. But options (and the data being tidy in the first place) make it easy to go your own way when you need to.
Variable names in models should be descriptive, not cryptic. This principle implies avoiding cryptic (and short) subscripts in favor of longer (but descriptive) ones. This is a matter of readability and accessibility of models to others. For example, a common pattern among Stan users (and in the Stan manual) is to use variables like
Jto refer to the number of elements in a group (e.g., number of participants) and a corresponding index likejto refer to specific elements in that group. I believe this sacrifices too much readability for the sake of concision; I prefer a pattern liken_participantfor the size of the group andparticipant(or a mnemonic short form likep) for specific elements. In functions where names are auto-generated (likecompose_data),tidybayeswill (by default) assume you want these sorts of more descriptive names; however, you can always override the default naming scheme.
Supported model types
tidybayes aims to support a variety of models with a
uniform interface. Currently supported models include rstan, cmdstanr, brms, rstanarm, runjags, rjags, jagsUI, coda::mcmc and
coda::mcmc.list, posterior::draws, MCMCglmm, and
anything with its own as.mcmc.list implementation. If you
install the tidybayes.rethinking
package, models from the rethinking package
are also supported.
For an up-to-date list of supported models, see
?"tidybayes-models".
Setup
The following libraries are required to run this vignette:
library(magrittr)
library(dplyr)
library(forcats)
library(modelr)
library(ggdist)
library(tidybayes)
library(ggplot2)
library(cowplot)
library(broom)
library(rstan)
library(rstanarm)
library(brms)
library(bayesplot)
library(RColorBrewer)
theme_set(theme_tidybayes() + panel_border())These options help Stan run faster:
rstan_options(auto_write = TRUE)
options(mc.cores = parallel::detectCores())Example dataset
To demonstrate tidybayes, we will use a simple dataset
with 10 observations from 5 conditions each:
set.seed(5)
n = 10
n_condition = 5
ABC =
tibble(
condition = factor(rep(c("A","B","C","D","E"), n)),
response = rnorm(n * 5, c(0,1,2,1,-1), 0.5)
)A snapshot of the data looks like this:
head(ABC, 10)## # A tibble: 10 × 2
## condition response
## <fct> <dbl>
## 1 A -0.420
## 2 B 1.69
## 3 C 1.37
## 4 D 1.04
## 5 E -0.144
## 6 A -0.301
## 7 B 0.764
## 8 C 1.68
## 9 D 0.857
## 10 E -0.931This is a typical tidy format data frame: one observation per row. Graphically:
ABC %>%
ggplot(aes(x = response, y = fct_rev(condition))) +
geom_point(alpha = 0.5) +
ylab("condition")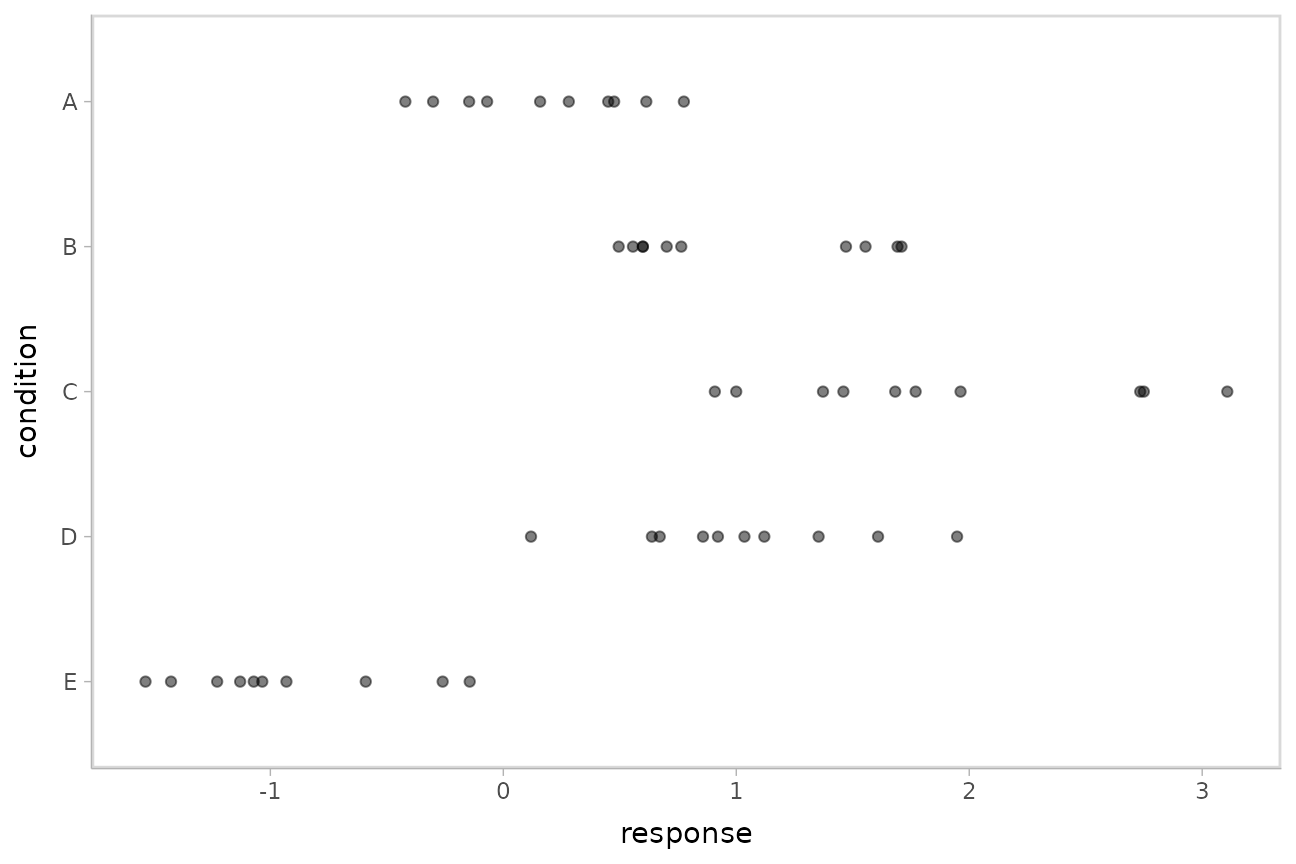
Using compose_data to prepare a data frame for the
model
Shunting data from a data frame into a format usable in samplers like
JAGS or Stan can involve a tedious set of operations, like generating
index variables storing the number of operations or the number of levels
in a factor. compose_data automates these operations.
A hierarchical model of our example data might fit an overall mean
across the conditions (overall_mean), the standard
deviation of the condition means (condition_mean_sd), the
mean within each condition (condition_mean[condition]) and
the standard deviation of the responses given a condition mean
(response_sd):
data {
int<lower=1> n;
int<lower=1> n_condition;
int<lower=1, upper=n_condition> condition[n];
real response[n];
}
parameters {
real overall_mean;
vector[n_condition] condition_zoffset;
real<lower=0> response_sd;
real<lower=0> condition_mean_sd;
}
transformed parameters {
vector[n_condition] condition_mean;
condition_mean = overall_mean + condition_zoffset * condition_mean_sd;
}
model {
response_sd ~ cauchy(0, 1); // => half-cauchy(0, 1)
condition_mean_sd ~ cauchy(0, 1); // => half-cauchy(0, 1)
overall_mean ~ normal(0, 5);
condition_zoffset ~ normal(0, 1); // => condition_mean ~ normal(overall_mean, condition_mean_sd)
for (i in 1:n) {
response[i] ~ normal(condition_mean[condition[i]], response_sd);
}
}We have compiled and loaded this model into the variable
ABC_stan.
This model expects these variables as input:
-
n: number of observations -
n_condition: number of conditions -
condition: a vector of integers indicating the condition of each observation -
response: a vector of observations
Our data frame (ABC) only has response and
condition, and condition is in the wrong
format (it is a factor instead of numeric). However,
compose_data can generate a list containing the above
variables in the correct format automatically. It recognizes that
condition is a factor and converts it to a numeric, adds
the n_condition variable automatically containing the
number of levels in condition, and adds the n
column containing the number of observations (number of rows in the data
frame):
compose_data(ABC)## $condition
## [1] 1 2 3 4 5 1 2 3 4 5 1 2 3 4 5 1 2 3 4 5 1 2 3 4 5 1 2 3 4 5 1 2 3 4 5 1 2 3 4 5 1 2 3 4 5 1 2 3 4 5
##
## $n_condition
## [1] 5
##
## $response
## [1] -0.42042774 1.69217967 1.37225407 1.03507138 -0.14427956 -0.30145399 0.76391681 1.68231434 0.85711318
## [10] -0.93094589 0.61381517 0.59911027 1.45980370 0.92123282 -1.53588002 -0.06949307 0.70134345 0.90801662
## [19] 1.12040863 -1.12967770 0.45025597 1.47093470 2.73398095 1.35338054 -0.59049553 -0.14674092 1.70929454
## [28] 2.74938691 0.67145895 -1.42639772 0.15795752 1.55484708 3.10773029 1.60855182 -0.26038911 0.47578692
## [37] 0.49523368 0.99976363 0.11890706 -1.07130406 0.77503018 0.59878841 1.96271054 1.94783398 -1.22828447
## [46] 0.28111168 0.55649574 1.76987771 0.63783576 -1.03460558
##
## $n
## [1] 50This makes it easy to skip right to running the model without munging the data yourself:
m = sampling(ABC_stan, data = compose_data(ABC), control = list(adapt_delta = 0.99))The results look like this:
m## Inference for Stan model: anon_model.
## 4 chains, each with iter=2000; warmup=1000; thin=1;
## post-warmup draws per chain=1000, total post-warmup draws=4000.
##
## mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
## overall_mean 0.63 0.02 0.62 -0.55 0.28 0.63 0.95 1.87 915 1
## condition_zoffset[1] -0.39 0.02 0.49 -1.41 -0.71 -0.39 -0.05 0.58 973 1
## condition_zoffset[2] 0.35 0.02 0.49 -0.62 0.04 0.33 0.66 1.31 902 1
## condition_zoffset[3] 1.11 0.02 0.59 -0.02 0.70 1.11 1.50 2.30 868 1
## condition_zoffset[4] 0.37 0.02 0.49 -0.62 0.04 0.36 0.69 1.31 905 1
## condition_zoffset[5] -1.39 0.02 0.65 -2.74 -1.81 -1.36 -0.94 -0.23 1031 1
## response_sd 0.56 0.00 0.06 0.46 0.52 0.56 0.60 0.70 1793 1
## condition_mean_sd 1.23 0.02 0.51 0.62 0.90 1.12 1.43 2.50 997 1
## condition_mean[1] 0.20 0.00 0.17 -0.14 0.09 0.20 0.32 0.55 4885 1
## condition_mean[2] 1.00 0.00 0.18 0.65 0.89 1.01 1.12 1.34 4606 1
## condition_mean[3] 1.84 0.00 0.18 1.48 1.72 1.84 1.95 2.19 4893 1
## condition_mean[4] 1.02 0.00 0.18 0.68 0.90 1.02 1.14 1.37 4274 1
## condition_mean[5] -0.89 0.00 0.18 -1.23 -1.01 -0.89 -0.77 -0.53 4725 1
## lp__ 0.30 0.08 2.34 -5.02 -1.08 0.63 2.06 3.79 915 1
##
## Samples were drawn using NUTS(diag_e) at Sat Sep 14 23:35:52 2024.
## For each parameter, n_eff is a crude measure of effective sample size,
## and Rhat is the potential scale reduction factor on split chains (at
## convergence, Rhat=1).Extracting draws from a fit in tidy-format using
spread_draws
Extracting model variable indices into a separate column in a tidy format data frame
Now that we have our results, the fun begins: getting the draws out in a tidy format! The default methods in Stan for extracting draws from the model do so in a nested format:
## List of 6
## $ overall_mean : num [1:4000(1d)] 1.113 0.818 0.432 0.794 0.528 ...
## ..- attr(*, "dimnames")=List of 1
## .. ..$ iterations: NULL
## $ condition_zoffset: num [1:4000, 1:5] -0.9652 -0.4328 -0.0276 -0.4277 -0.0597 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ iterations: NULL
## .. ..$ : NULL
## $ response_sd : num [1:4000(1d)] 0.696 0.587 0.651 0.571 0.566 ...
## ..- attr(*, "dimnames")=List of 1
## .. ..$ iterations: NULL
## $ condition_mean_sd: num [1:4000(1d)] 0.974 1.068 0.53 1.456 0.951 ...
## ..- attr(*, "dimnames")=List of 1
## .. ..$ iterations: NULL
## $ condition_mean : num [1:4000, 1:5] 0.173 0.356 0.417 0.171 0.472 ...
## ..- attr(*, "dimnames")=List of 2
## .. ..$ iterations: NULL
## .. ..$ : NULL
## $ lp__ : num [1:4000(1d)] -1.598 0.887 -5.352 3.171 0.319 ...
## ..- attr(*, "dimnames")=List of 1
## .. ..$ iterations: NULLThere are also methods for extracting draws as matrices or data frames in Stan (and other model types, such as JAGS and MCMCglmm, have their own formats).
The spread_draws method yields a common format for all
model types supported by tidybayes. It lets us instead
extract draws into a data frame in tidy format, with a
.chain and .iteration column storing the chain
and iteration for each row (if available), a .draw column
that uniquely indexes each draw, and the remaining columns corresponding
to model variables or variable indices. The spread_draws
method accepts any number of column specifications, which can include
names for variables and names for variable indices. For example, we can
extract the condition_mean variable as a tidy data frame,
and put the value of its first (and only) index into the
condition column, using a syntax that directly echoes how
we would specify indices of the condition_mean variable in
the model itself:
m %>%
spread_draws(condition_mean[condition]) %>%
head(10)## # A tibble: 10 × 5
## # Groups: condition [1]
## condition condition_mean .chain .iteration .draw
## <int> <dbl> <int> <int> <int>
## 1 1 0.00544 1 1 1
## 2 1 -0.0836 1 2 2
## 3 1 0.0324 1 3 3
## 4 1 0.113 1 4 4
## 5 1 0.157 1 5 5
## 6 1 0.218 1 6 6
## 7 1 0.276 1 7 7
## 8 1 0.0130 1 8 8
## 9 1 0.152 1 9 9
## 10 1 0.192 1 10 10Automatically converting columns and indices back into their original data types
As-is, the resulting variables don’t know anything about where their
indices came from. The index of the condition_mean variable
was originally derived from the condition factor in the
ABC data frame. But Stan doesn’t know this: it is just a
numeric index to Stan, so the condition column just
contains numbers (1, 2, 3, 4, 5) instead of the factor
levels these numbers correspond to
("A", "B", "C", "D", "E").
We can recover this missing type information by passing the model
through recover_types before using
spread_draws. In itself recover_types just
returns a copy of the model, with some additional attributes that store
the type information from the data frame (or other objects) that you
pass to it. This doesn’t have any useful effect by itself, but functions
like spread_draws use this information to convert any
column or index back into the data type of the column with the same name
in the original data frame. In this example, spread_draws
recognizes that the condition column was a factor with five
levels ("A", "B", "C", "D", "E") in the original data
frame, and automatically converts it back into a factor:
m %>%
recover_types(ABC) %>%
spread_draws(condition_mean[condition]) %>%
head(10)## # A tibble: 10 × 5
## # Groups: condition [1]
## condition condition_mean .chain .iteration .draw
## <fct> <dbl> <int> <int> <int>
## 1 A 0.00544 1 1 1
## 2 A -0.0836 1 2 2
## 3 A 0.0324 1 3 3
## 4 A 0.113 1 4 4
## 5 A 0.157 1 5 5
## 6 A 0.218 1 6 6
## 7 A 0.276 1 7 7
## 8 A 0.0130 1 8 8
## 9 A 0.152 1 9 9
## 10 A 0.192 1 10 10Because we often want to make multiple separate calls to
spread_draws, it is often convenient to decorate the
original model using recover_types immediately after it has
been fit, so we only have to call it once:
m %<>% recover_types(ABC)Now we can omit the recover_types call before subsequent
calls to spread_draws.
Point summaries and intervals with the point_interval
functions: [median|mean|mode]_[qi|hdi]
With simple variables, wide format
tidybayes provides a family of functions for generating
point summaries and intervals from draws in a tidy format. These
functions follow the naming scheme
[median|mean|mode]_[qi|hdi], for example,
median_qi, mean_qi, mode_hdi, and
so on. The first name (before the _) indicates the type of
point summary, and the second name indicates the type of interval.
qi yields a quantile interval (a.k.a. equi-tailed interval,
central interval, or percentile interval) and hdi yields a
highest density interval. Custom point or interval functions can also be
applied using the point_interval function.
For example, we might extract the draws corresponding to the overall mean and standard deviation of observations:
m %>%
spread_draws(overall_mean, response_sd) %>%
head(10)## # A tibble: 10 × 5
## .chain .iteration .draw overall_mean response_sd
## <int> <int> <int> <dbl> <dbl>
## 1 1 1 1 0.0672 0.576
## 2 1 2 2 0.0361 0.576
## 3 1 3 3 1.17 0.551
## 4 1 4 4 0.378 0.576
## 5 1 5 5 0.359 0.583
## 6 1 6 6 0.332 0.621
## 7 1 7 7 0.226 0.641
## 8 1 8 8 0.107 0.637
## 9 1 9 9 0.225 0.609
## 10 1 10 10 -0.0395 0.521Like with condition_mean[condition], this gives us a
tidy data frame. If we want the median and 95% quantile interval of the
variables, we can apply median_qi:
m %>%
spread_draws(overall_mean, response_sd) %>%
median_qi(overall_mean, response_sd)## overall_mean overall_mean.lower overall_mean.upper response_sd response_sd.lower response_sd.upper .width .point
## 1 0.6331377 -0.5458905 1.868912 0.5575944 0.455513 0.6976528 0.95 median
## .interval
## 1 qimedian_qi summarizes each input column using its median.
If there are multiple columns to summarize, each gets its own
x.lower and x.upper column (for each column
x) corresponding to the bounds of the .width%
interval. If there is only one column, the names .lower and
.upper are used for the interval bounds.
We can specify the columns we want to get medians and intervals from,
as above, or if we omit the list of columns, median_qi will
use every column that is not a grouping column or a special column (like
.chain, .iteration, or .draw).
Thus in the above example, overall_mean and
response_sd are redundant arguments to
median_qi because they are also the only columns we
gathered from the model. So we can simplify the previous code to the
following:
m %>%
spread_draws(overall_mean, response_sd) %>%
median_qi()## overall_mean overall_mean.lower overall_mean.upper response_sd response_sd.lower response_sd.upper .width .point
## 1 0.6331377 -0.5458905 1.868912 0.5575944 0.455513 0.6976528 0.95 median
## .interval
## 1 qiWith indexed variables
When we have a variable with one or more indices, such as
condition_mean, we can apply median_qi (or
other functions in the point_interval family) as we did
before:
m %>%
spread_draws(condition_mean[condition]) %>%
median_qi()## # A tibble: 5 × 7
## condition condition_mean .lower .upper .width .point .interval
## <fct> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 A 0.199 -0.142 0.549 0.95 median qi
## 2 B 1.01 0.651 1.34 0.95 median qi
## 3 C 1.84 1.48 2.19 0.95 median qi
## 4 D 1.02 0.681 1.37 0.95 median qi
## 5 E -0.890 -1.23 -0.529 0.95 median qiHow did median_qi know what to aggregate? Data frames
returned by spread_draws are automatically grouped by all
index variables you pass to it; in this case, that means it groups by
condition. median_qi respects groups, and
calculates the point summaries and intervals within all groups. Then,
because no columns were passed to median_qi, it acts on the
only non-special (.-prefixed) and non-group column,
condition_mean. So the above shortened syntax is equivalent
to this more verbose call:
m %>%
spread_draws(condition_mean[condition]) %>%
group_by(condition) %>% # this line not necessary (done automatically by spread_draws)
median_qi(condition_mean)## # A tibble: 5 × 7
## condition condition_mean .lower .upper .width .point .interval
## <fct> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 A 0.199 -0.142 0.549 0.95 median qi
## 2 B 1.01 0.651 1.34 0.95 median qi
## 3 C 1.84 1.48 2.19 0.95 median qi
## 4 D 1.02 0.681 1.37 0.95 median qi
## 5 E -0.890 -1.23 -0.529 0.95 median qiWhen given only a single column, median_qi will use the
names .lower and .upper for the lower and
upper ends of the intervals.
tidybayes also provides an implementation of
posterior::summarise_draws() for grouped data frames
(tidybayes::summaries_draws.grouped_df()), which you can
use to quickly get convergence diagnostics:
m %>%
spread_draws(condition_mean[condition]) %>%
summarise_draws()## # A tibble: 5 × 11
## # Groups: condition [5]
## condition variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
## <fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 A condition_mean 0.201 0.199 0.172 0.171 -0.0847 0.481 1.00 4779. 3172.
## 2 B condition_mean 1.00 1.01 0.176 0.173 0.711 1.28 1.00 4660. 3492.
## 3 C condition_mean 1.84 1.84 0.178 0.174 1.54 2.12 1.00 4794. 3601.
## 4 D condition_mean 1.02 1.02 0.177 0.178 0.738 1.31 1.00 4303. 3060.
## 5 E condition_mean -0.888 -0.890 0.180 0.177 -1.18 -0.593 1.00 4758. 3248.Plotting points and intervals
Using geom_pointinterval
Plotting medians and intervals is straightforward using
ggdist::geom_pointinterval() or
ggdist::stat_pointinterval(), which are similar to
ggplot2::geom_pointrange() but with sensible defaults for
multiple intervals. For example:
m %>%
spread_draws(condition_mean[condition]) %>%
ggplot(aes(y = fct_rev(condition), x = condition_mean)) +
stat_pointinterval()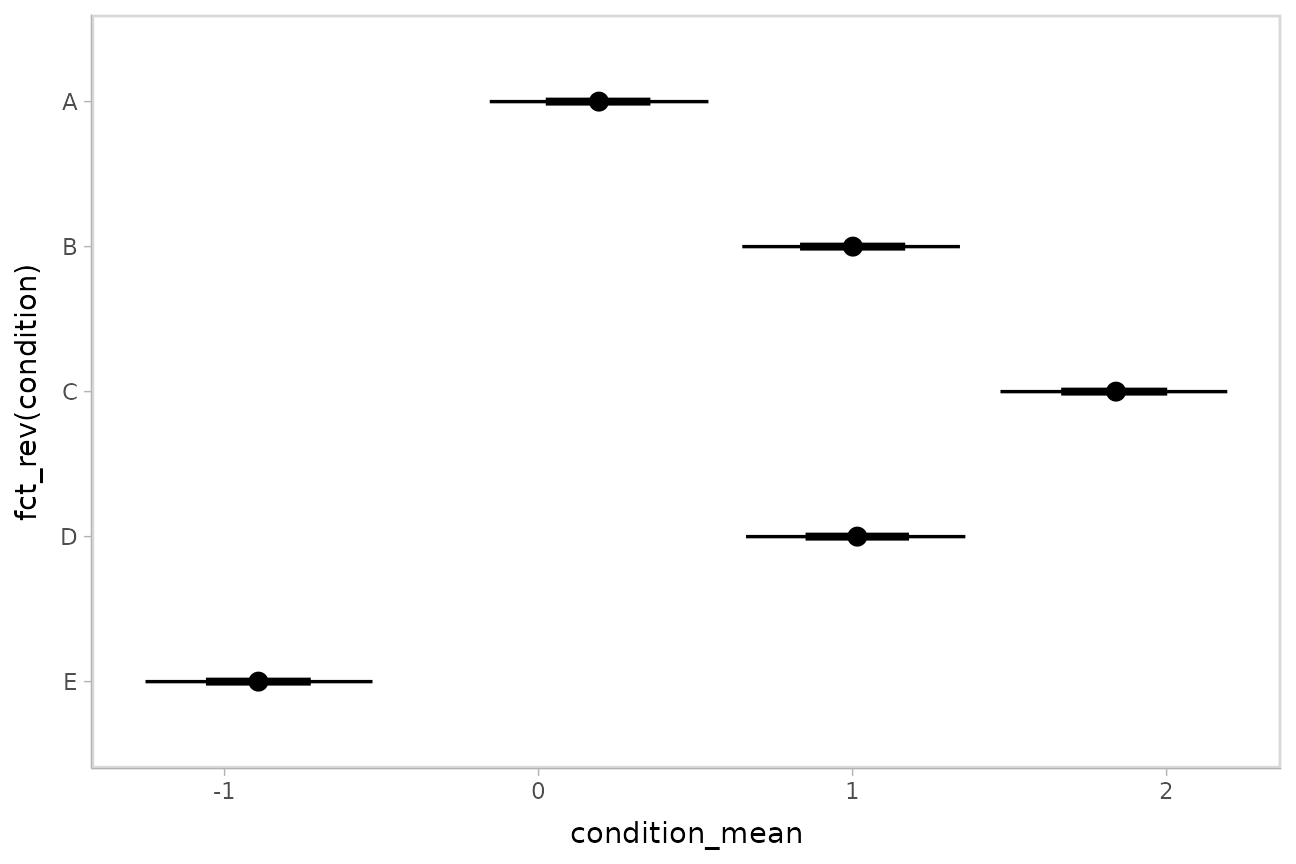
These functions have .width = c(.66, .95) by default
(showing 66% and 95% intervals), but this can be changed by passing a
.width argument to
ggdist::stat_pointinterval().
Intervals with posterior violins (“eye plots”):
stat_eye
The ggdist::stat_halfeye() geom provides a shortcut to
generating “half-eye plots” (combinations of intervals and densities).
This example also demonstrates how to change the interval probability
(here, to 90% and 50% intervals):
m %>%
spread_draws(condition_mean[condition]) %>%
ggplot(aes(y = fct_rev(condition), x = condition_mean)) +
stat_halfeye(.width = c(.90, .5))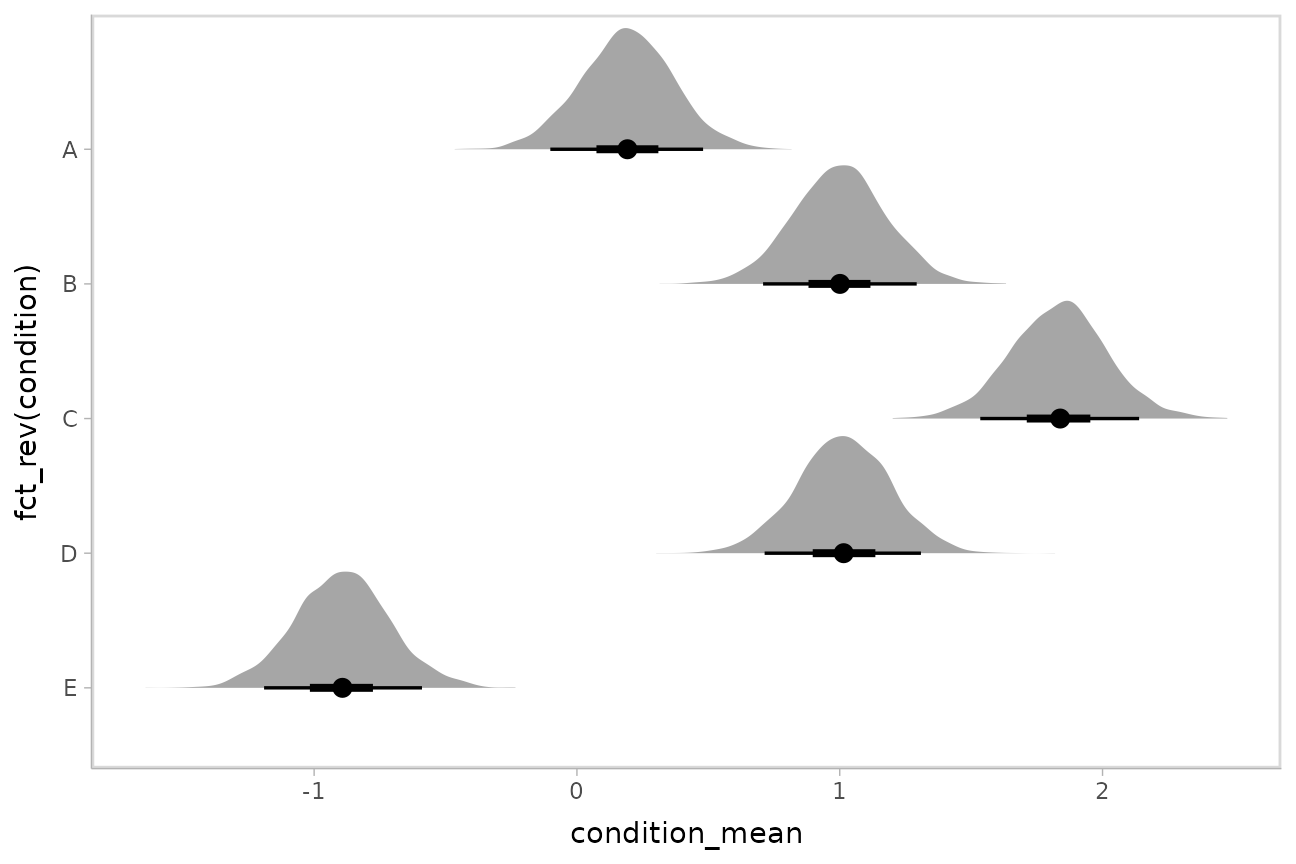
Or say you want to annotate portions of the densities in color; the
fill aesthetic can vary within a slab in all geoms and
stats in the ggdist::geom_slabinterval() family, including
ggdist::stat_halfeye(). For example, if you want to
annotate a domain-specific region of practical equivalence (ROPE), you
could do something like this:
m %>%
spread_draws(condition_mean[condition]) %>%
ggplot(aes(y = fct_rev(condition), x = condition_mean, fill = after_stat(abs(x) < .8))) +
stat_halfeye() +
geom_vline(xintercept = c(-.8, .8), linetype = "dashed") +
scale_fill_manual(values = c("gray80", "skyblue"))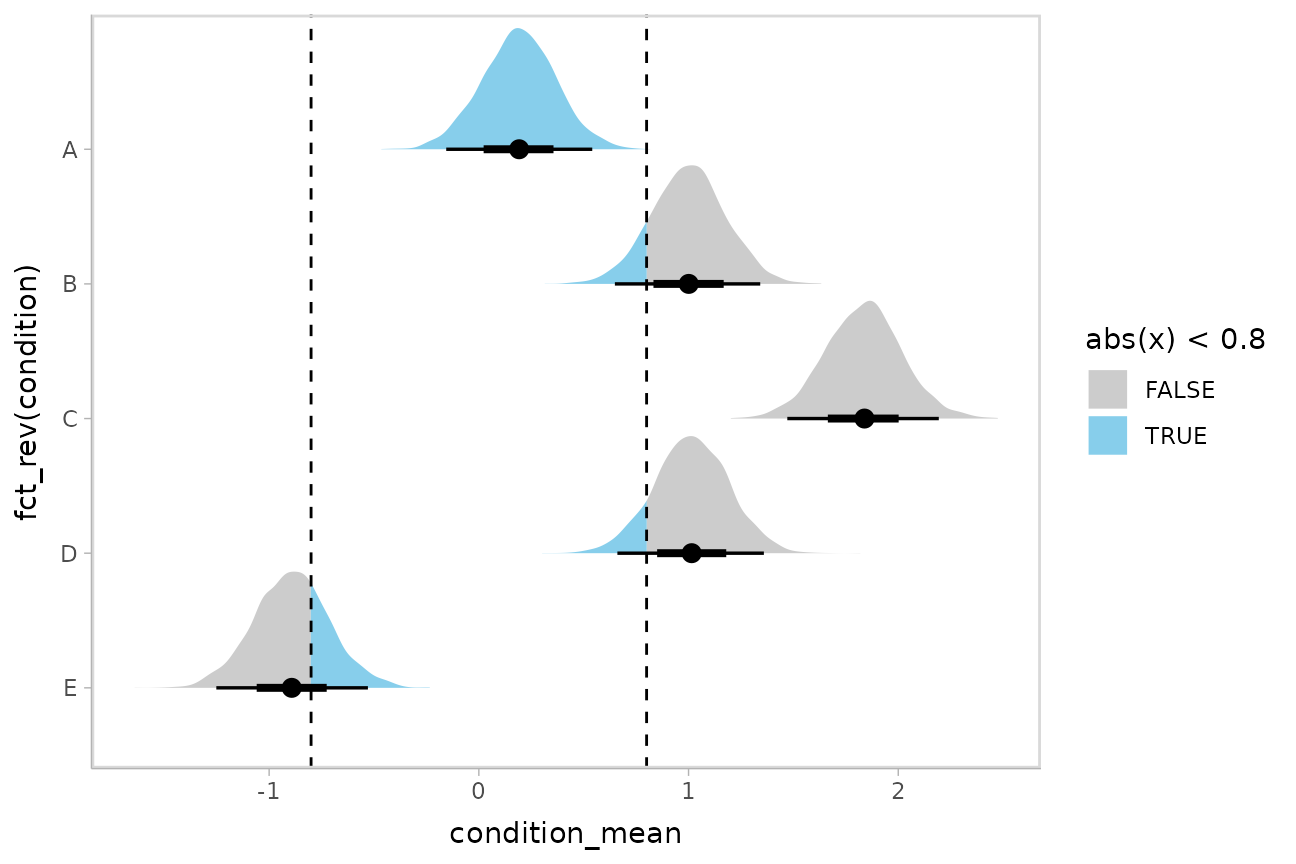
Other visualizations of distributions:
stat_slabinterval
There are a variety of additional stats for visualizing distributions
in the ggdist::geom_slabinterval() family of stats and
geoms:
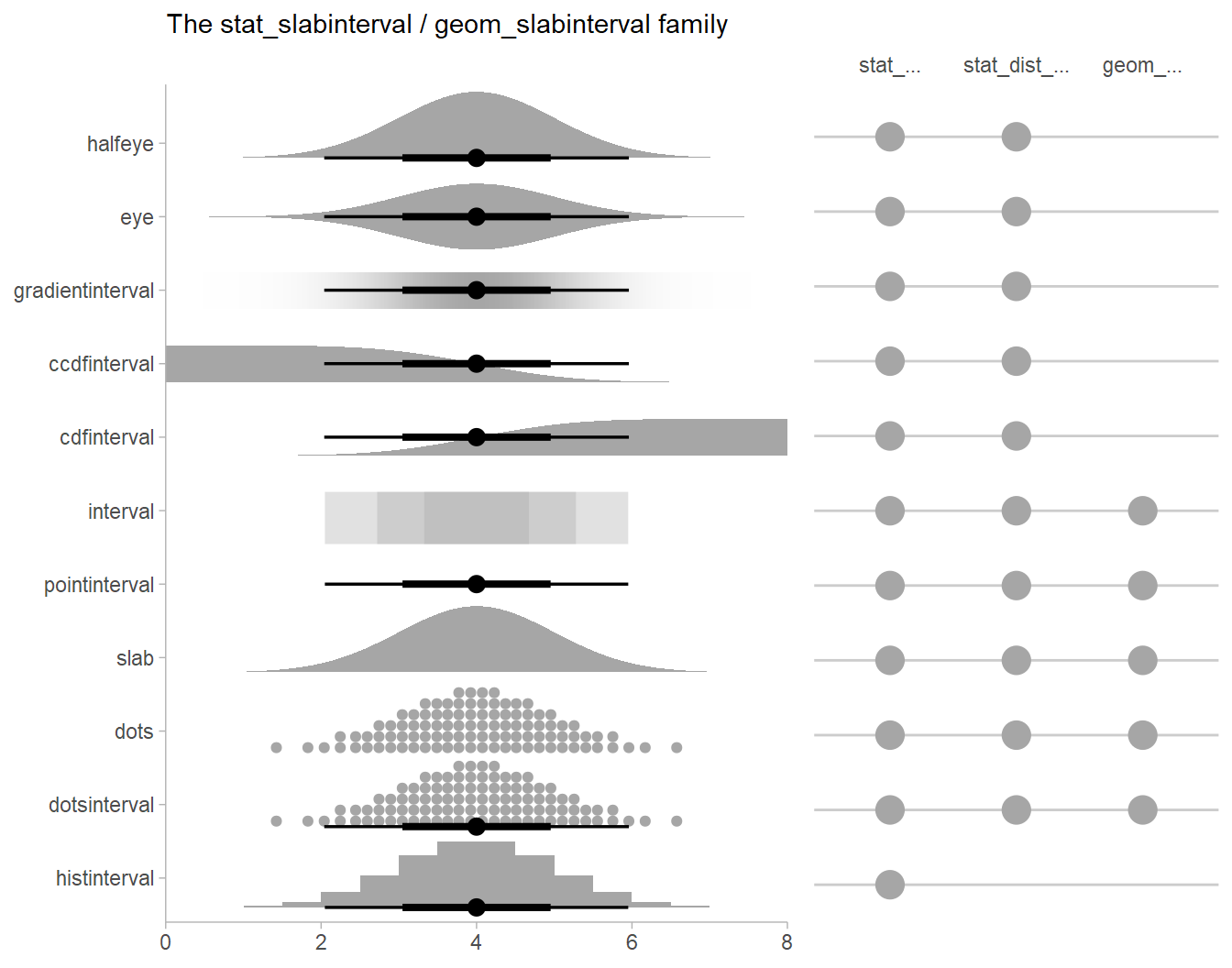
See vignette("slabinterval", package = "ggdist") for an
overview.
Intervals with multiple probability levels: the
.width = argument
If you wish to summarise the data before plotting (sometimes useful
for large samples), median_qi() and its sister functions
can also produce an arbitrary number of probability intervals by setting
the .width = argument:
m %>%
spread_draws(condition_mean[condition]) %>%
median_qi(.width = c(.95, .8, .5))## # A tibble: 15 × 7
## condition condition_mean .lower .upper .width .point .interval
## <fct> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 A 0.199 -0.142 0.549 0.95 median qi
## 2 B 1.01 0.651 1.34 0.95 median qi
## 3 C 1.84 1.48 2.19 0.95 median qi
## 4 D 1.02 0.681 1.37 0.95 median qi
## 5 E -0.890 -1.23 -0.529 0.95 median qi
## 6 A 0.199 -0.0145 0.421 0.8 median qi
## 7 B 1.01 0.777 1.22 0.8 median qi
## 8 C 1.84 1.61 2.06 0.8 median qi
## 9 D 1.02 0.797 1.25 0.8 median qi
## 10 E -0.890 -1.11 -0.658 0.8 median qi
## 11 A 0.199 0.0870 0.317 0.5 median qi
## 12 B 1.01 0.887 1.12 0.5 median qi
## 13 C 1.84 1.72 1.95 0.5 median qi
## 14 D 1.02 0.904 1.14 0.5 median qi
## 15 E -0.890 -1.01 -0.765 0.5 median qiThe results are in a tidy format: one row per index
(condition) and probability level (.width).
This facilitates plotting. For example, assigning -.width
to the linewidth aesthetic will show all intervals, making
thicker lines correspond to smaller intervals:
m %>%
spread_draws(condition_mean[condition]) %>%
median_qi(.width = c(.95, .66)) %>%
ggplot(aes(
y = fct_rev(condition), x = condition_mean, xmin = .lower, xmax = .upper,
# size = -.width means smaller probability interval => thicker line
# this can be omitted, geom_pointinterval includes it automatically
# if a .width column is in the input data.
linewidth = -.width
)) +
geom_pointinterval()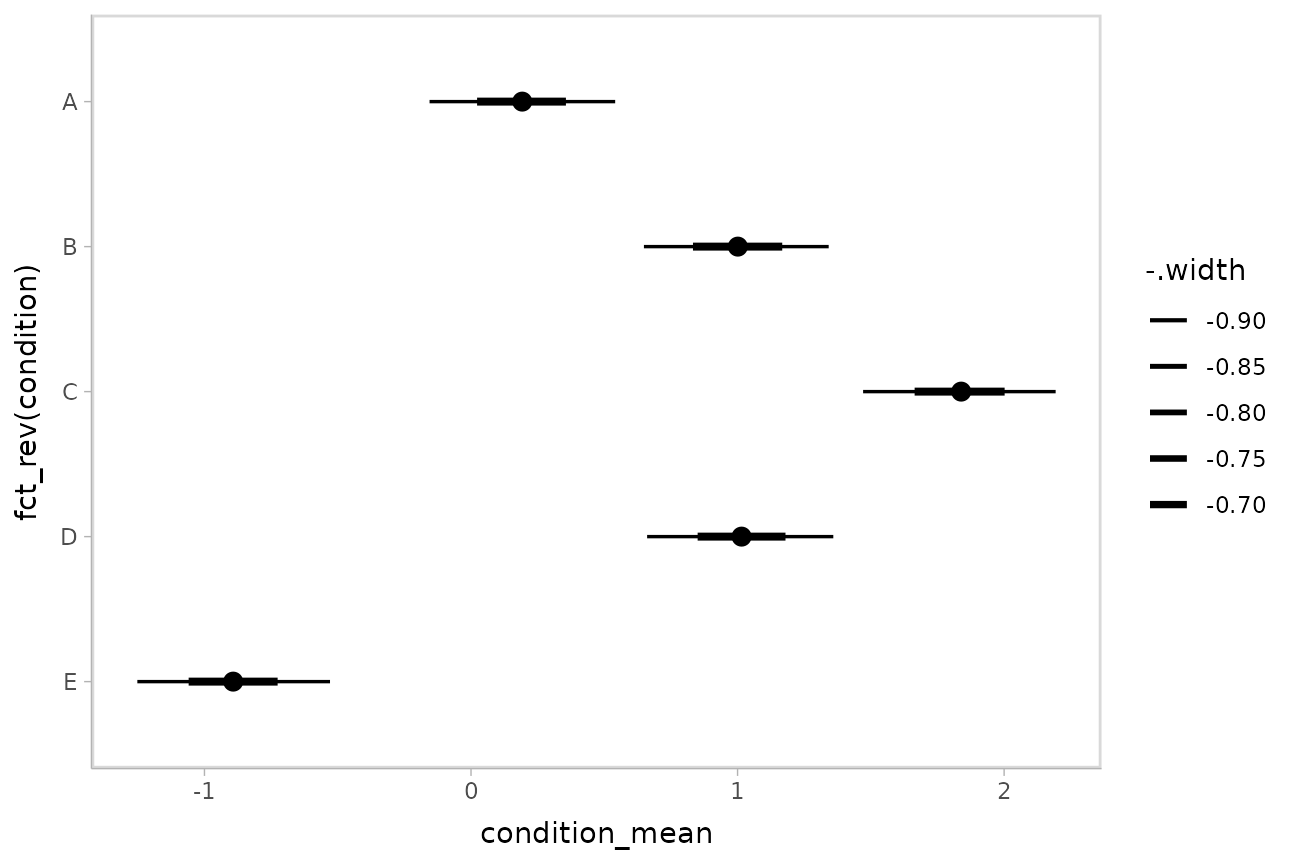
ggdist::geom_pointinterval() includes
size = -.width as a default aesthetic mapping to facilitate
exactly this usage.
Plotting posteriors as quantile dotplots
Intervals are nice if the alpha level happens to line up with whatever decision you are trying to make, but getting a shape of the posterior is better (hence eye plots, above). On the other hand, making inferences from density plots is imprecise (estimating the area of one shape as a proportion of another is a hard perceptual task). Reasoning about probability in frequency formats is easier, motivating quantile dotplots (Kay et al. 2016, Fernandes et al. 2018), which also allow precise estimation of arbitrary intervals (down to the dot resolution of the plot, 100 in the example below).
Within the slabinterval family of geoms in tidybayes is the
dots and dotsinterval family, which
automatically determine appropriate bin sizes for dotplots and can
calculate quantiles from samples to construct quantile dotplots.
ggdist::stat_dots() is the variant designed for use on
samples:
m %>%
spread_draws(condition_mean[condition]) %>%
ggplot(aes(x = condition_mean, y = fct_rev(condition))) +
stat_dotsinterval(quantiles = 100)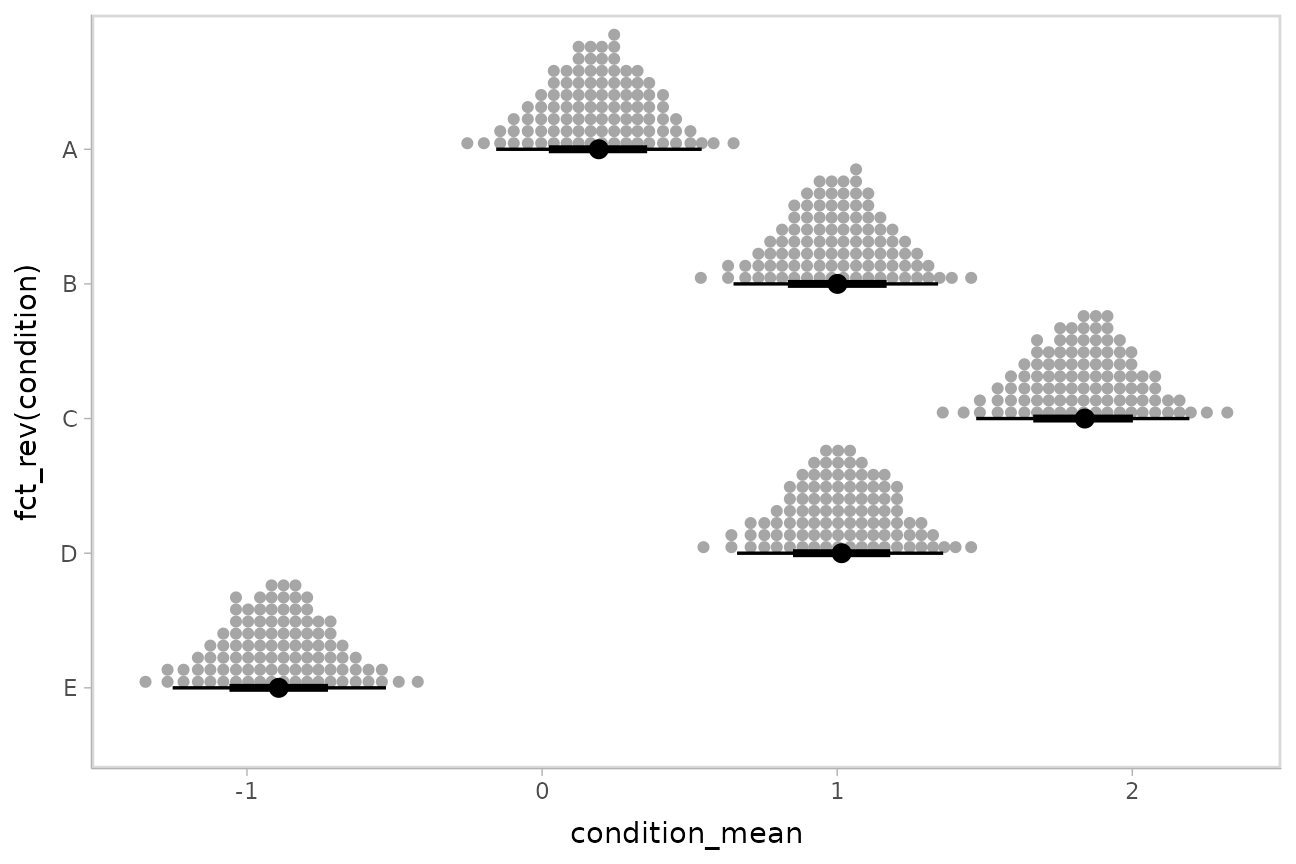
The idea is to get away from thinking about the posterior as indicating one canonical point or interval, but instead to represent it as (say) 100 approximately equally likely points.
Alternative point summaries and intervals: median, mean, mode; qi, hdi, hdci
The point_interval() family of functions follow the
naming scheme [median|mean|mode]_[qi|hdi|hdci], and all
work in the same way as median_qi(): they take a series of
names (or expressions calculated on columns) and summarize those columns
with the corresponding point summary function (median, mean, or mode)
and interval (qi, hdi, or hdci). qi yields a quantile
interval (a.k.a. equi-tailed interval, central interval, or percentile
interval), hdi yields one or more highest (posterior)
density interval(s), and hdci yields a single (possibly)
highest-density continuous interval. These can be used in any
combination desired.
The *_hdi functions have an additional difference: In
the case of multimodal distributions, they may return multiple intervals
for each probability level. Here are some draws from a multimodal normal
mixture:
Passed through mode_hdi(), we get multiple intervals at
the 80% probability level:
## # A tibble: 2 × 6
## x .lower .upper .width .point .interval
## <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 -0.0605 -1.46 1.54 0.8 mode hdi
## 2 -0.0605 3.11 5.01 0.8 mode hdiThis is easier to see when plotted:
multimodal_draws %>%
ggplot(aes(x = x)) +
stat_slab(aes(y = 0)) +
stat_pointinterval(aes(y = -0.5), point_interval = median_qi, .width = c(.95, .80)) +
annotate("text", label = "median, 80% and 95% quantile intervals", x = 6, y = -0.5, hjust = 0, vjust = 0.3) +
stat_pointinterval(aes(y = -0.25), point_interval = mode_hdi, .width = c(.95, .80)) +
annotate("text", label = "mode, 80% and 95% highest-density intervals", x = 6, y = -0.25, hjust = 0, vjust = 0.3) +
xlim(-3.25, 18) +
scale_y_continuous(breaks = NULL)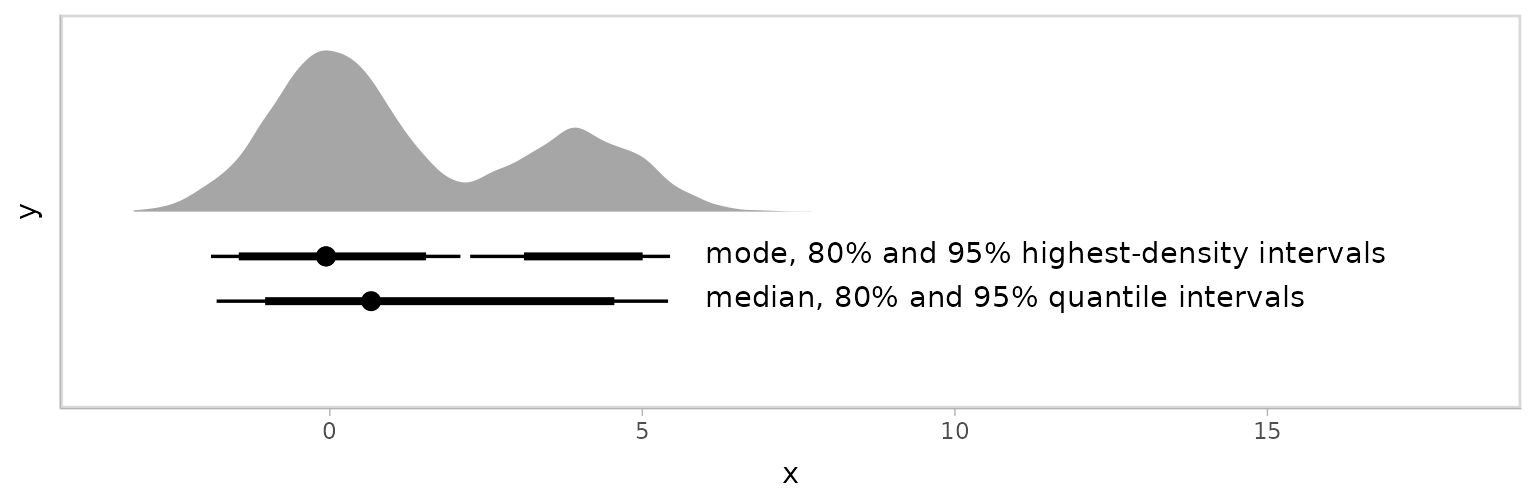
Combining variables with different indices in a single tidy format data frame
spread_draws() supports extracting variables that have
different indices. It automatically matches up indices with the same
name, and duplicates values as necessary to produce one row per all
combination of levels of all indices. For example, we might want to
calculate the difference between each condition mean and the overall
mean. To do that, we can extract draws from the overall mean and all
condition means:
m %>%
spread_draws(overall_mean, condition_mean[condition]) %>%
head(10)## # A tibble: 10 × 6
## # Groups: condition [5]
## .chain .iteration .draw overall_mean condition condition_mean
## <int> <int> <int> <dbl> <fct> <dbl>
## 1 1 1 1 0.0672 A 0.00544
## 2 1 1 1 0.0672 B 1.03
## 3 1 1 1 0.0672 C 1.84
## 4 1 1 1 0.0672 D 1.25
## 5 1 1 1 0.0672 E -0.724
## 6 1 2 2 0.0361 A -0.0836
## 7 1 2 2 0.0361 B 0.873
## 8 1 2 2 0.0361 C 1.78
## 9 1 2 2 0.0361 D 1.12
## 10 1 2 2 0.0361 E -0.884Within each draw, overall_mean is repeated as necessary
to correspond to every index of condition_mean. Thus, the
dplyr::mutate() function can be used to take the
differences over all rows, then we can summarize with
median_qi():
m %>%
spread_draws(overall_mean, condition_mean[condition]) %>%
mutate(condition_offset = condition_mean - overall_mean) %>%
median_qi(condition_offset)## # A tibble: 5 × 7
## condition condition_offset .lower .upper .width .point .interval
## <fct> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 A -0.420 -1.69 0.754 0.95 median qi
## 2 B 0.363 -0.863 1.59 0.95 median qi
## 3 C 1.20 -0.0333 2.44 0.95 median qi
## 4 D 0.397 -0.885 1.61 0.95 median qi
## 5 E -1.50 -2.85 -0.340 0.95 median qiPosterior predictions
We can use combinations of variables with difference indices to
generate predictions from the model. In this case, we can combine the
condition means with the residual standard deviation to generate
predictive distributions from the model, then show the distributions
using ggdist::stat_interval() and compare them to the
data:
m %>%
spread_draws(condition_mean[condition], response_sd) %>%
mutate(y_rep = rnorm(n(), condition_mean, response_sd)) %>%
median_qi(y_rep, .width = c(.95, .8, .5)) %>%
ggplot(aes(y = fct_rev(condition), x = y_rep)) +
geom_interval(aes(xmin = .lower, xmax = .upper)) + #auto-sets aes(color = fct_rev(ordered(.width)))
geom_point(aes(x = response), data = ABC) +
scale_color_brewer()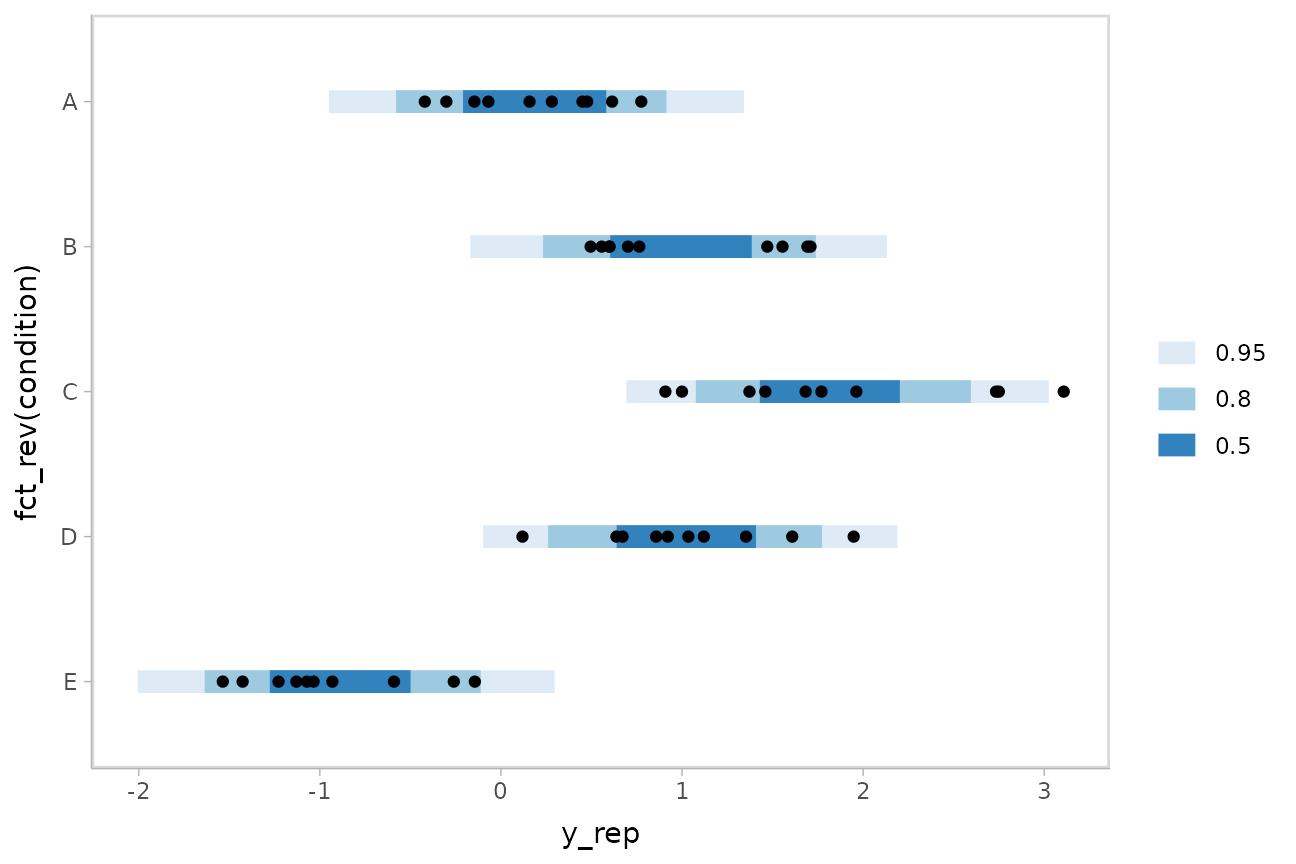
If this model is well-calibrated, about 95% of the data should be within the outer intervals, 80% in the next-smallest intervals, and 50% in the smallest intervals.
Posterior predictions with posterior distributions of means
Altogether, data, posterior predictions, and posterior distributions of the means:
draws = m %>%
spread_draws(condition_mean[condition], response_sd)
reps = draws %>%
mutate(y_rep = rnorm(n(), condition_mean, response_sd))
ABC %>%
ggplot(aes(y = condition)) +
stat_interval(aes(x = y_rep), .width = c(.95, .8, .5), data = reps) +
stat_pointinterval(aes(x = condition_mean), .width = c(.95, .66), position = position_nudge(y = -0.3), data = draws) +
geom_point(aes(x = response)) +
scale_color_brewer()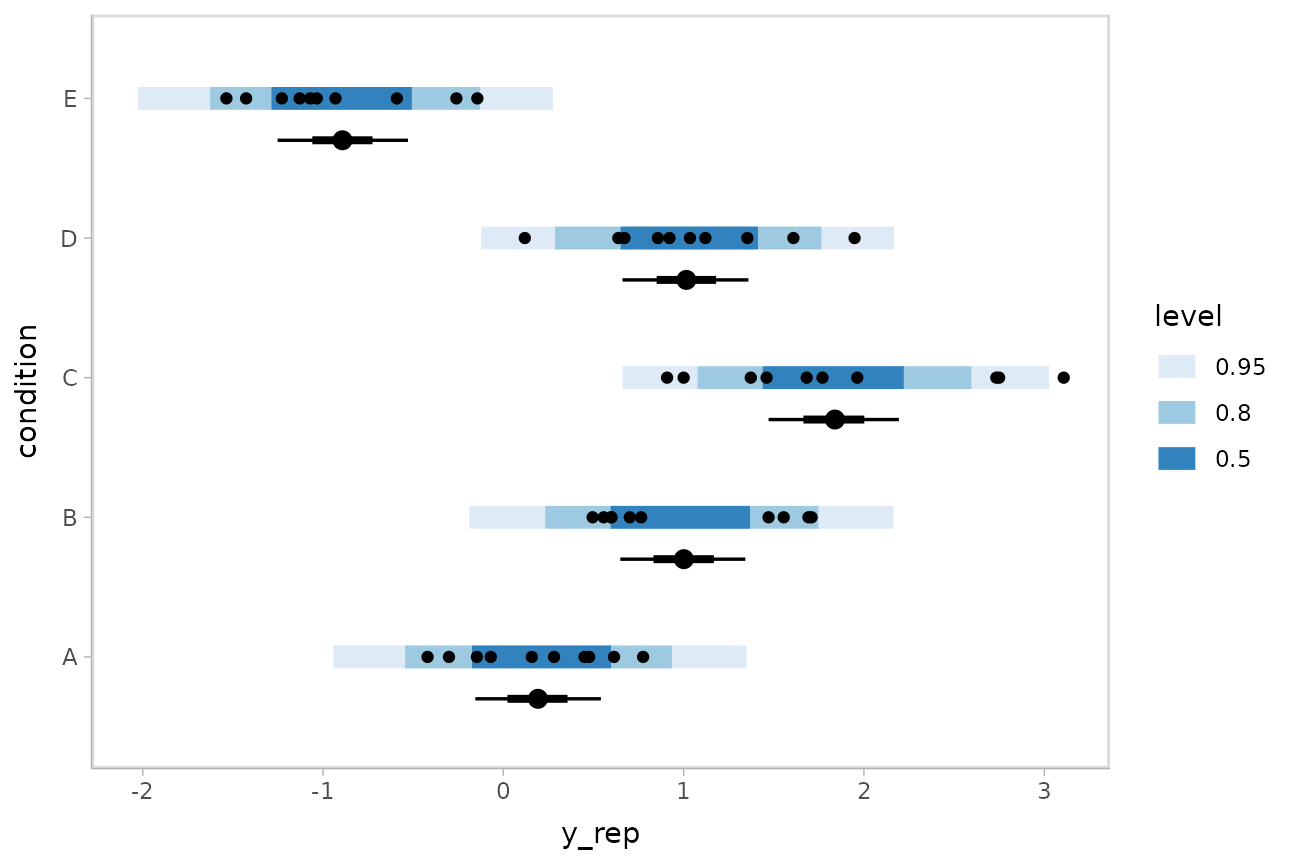
Comparing levels of a factor
compare_levels() allows us to compare the value of some
variable across levels of some factor. By default it computes all
pairwise differences, though this can be changed using the
comparison = argument:
m %>%
spread_draws(condition_mean[condition]) %>%
compare_levels(condition_mean, by = condition) %>%
ggplot(aes(y = condition, x = condition_mean)) +
stat_halfeye()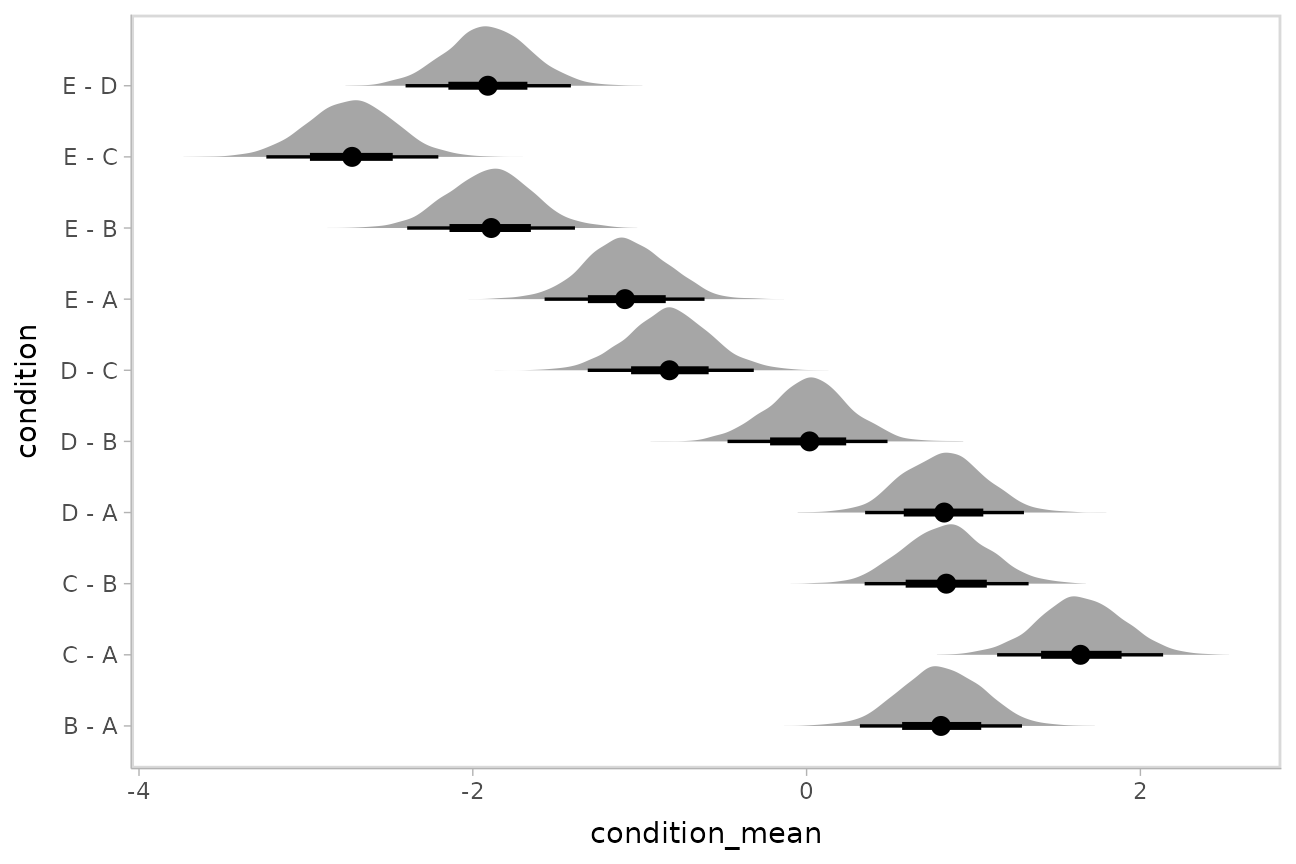
Gathering all model variable names into a single column:
gather_draws and gather_variables
We might also prefer all model variable names to be in a single
column (long-format) instead of as column names. There are two methods
for obtaining long-format data frames with tidybayes, whose
use depends on where and how in the data processing chain you might want
to transform into long-format: gather_draws() and
gather_variables(). There are also two methods for wide (or
semi-wide) format data frame, spread_draws() (described
previously) and tidy_draws().
gather_draws() is the counterpart to
spread_draws(), except it puts all variable names in a
.variable column and all draws in a .value
column:
m %>%
gather_draws(overall_mean, condition_mean[condition]) %>%
median_qi()## # A tibble: 6 × 8
## .variable condition .value .lower .upper .width .point .interval
## <chr> <fct> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 condition_mean A 0.199 -0.142 0.549 0.95 median qi
## 2 condition_mean B 1.01 0.651 1.34 0.95 median qi
## 3 condition_mean C 1.84 1.48 2.19 0.95 median qi
## 4 condition_mean D 1.02 0.681 1.37 0.95 median qi
## 5 condition_mean E -0.890 -1.23 -0.529 0.95 median qi
## 6 overall_mean NA 0.633 -0.546 1.87 0.95 median qiNote that condition = NA for the
overall_mean row, because it does not have an index with
that name in the specification passed to
gather_draws().
While this works well if we do not need to perform computations that
involve multiple columns, the semi-wide format returned by
spread_draws() is very useful for computations that involve
multiple columns names, such as the calculation of the
condition_offset above. If we want to make intermediate
computations on the format returned by spread_draws and
then gather variables into one column, we can use
gather_variables(), which will gather all non-grouped
variables that are not special columns (like .chain,
.iteration, or .draw):
m %>%
spread_draws(overall_mean, condition_mean[condition]) %>%
mutate(condition_offset = condition_mean - overall_mean) %>%
gather_variables() %>%
median_qi()## # A tibble: 15 × 8
## condition .variable .value .lower .upper .width .point .interval
## <fct> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 A condition_mean 0.199 -0.142 0.549 0.95 median qi
## 2 A condition_offset -0.420 -1.69 0.754 0.95 median qi
## 3 A overall_mean 0.633 -0.546 1.87 0.95 median qi
## 4 B condition_mean 1.01 0.651 1.34 0.95 median qi
## 5 B condition_offset 0.363 -0.863 1.59 0.95 median qi
## 6 B overall_mean 0.633 -0.546 1.87 0.95 median qi
## 7 C condition_mean 1.84 1.48 2.19 0.95 median qi
## 8 C condition_offset 1.20 -0.0333 2.44 0.95 median qi
## 9 C overall_mean 0.633 -0.546 1.87 0.95 median qi
## 10 D condition_mean 1.02 0.681 1.37 0.95 median qi
## 11 D condition_offset 0.397 -0.885 1.61 0.95 median qi
## 12 D overall_mean 0.633 -0.546 1.87 0.95 median qi
## 13 E condition_mean -0.890 -1.23 -0.529 0.95 median qi
## 14 E condition_offset -1.50 -2.85 -0.340 0.95 median qi
## 15 E overall_mean 0.633 -0.546 1.87 0.95 median qiNote how overall_mean is now repeated here for each
condition, because we have performed the gather after spreading model
variables across columns.
Finally, if we want raw model variable names as columns names instead
of having indices split out as their own column names, we can use
tidy_draws(). Generally speaking
spread_draws() and gather_draws() are
typically more useful than tidy_draws(), but it is provided
as a common method for generating data frames from many types of
Bayesian models, and is used internally by gather_draws()
and spread_draws():
m %>%
tidy_draws() %>%
head(10)## # A tibble: 10 × 23
## .chain .iteration .draw overall_mean `condition_zoffset[1]` `condition_zoffset[2]` `condition_zoffset[3]`
## <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
## 1 1 1 1 0.0672 -0.0237 0.369 0.682
## 2 1 2 2 0.0361 -0.0468 0.327 0.681
## 3 1 3 3 1.17 -0.827 -0.156 0.454
## 4 1 4 4 0.378 -0.260 0.603 1.66
## 5 1 5 5 0.359 -0.207 0.483 1.78
## 6 1 6 6 0.332 -0.132 0.563 1.89
## 7 1 7 7 0.226 0.0642 1.11 1.54
## 8 1 8 8 0.107 -0.0823 0.737 1.32
## 9 1 9 9 0.225 -0.0611 0.571 1.44
## 10 1 10 10 -0.0395 0.200 0.885 1.69
## # ℹ 16 more variables: `condition_zoffset[4]` <dbl>, `condition_zoffset[5]` <dbl>, response_sd <dbl>,
## # condition_mean_sd <dbl>, `condition_mean[1]` <dbl>, `condition_mean[2]` <dbl>, `condition_mean[3]` <dbl>,
## # `condition_mean[4]` <dbl>, `condition_mean[5]` <dbl>, lp__ <dbl>, accept_stat__ <dbl>, stepsize__ <dbl>,
## # treedepth__ <dbl>, n_leapfrog__ <dbl>, divergent__ <dbl>, energy__ <dbl>Combining tidy_draws() with
gather_variables() also allows us to derive similar output
to ggmcmc::ggs(), if desired:
m %>%
tidy_draws() %>%
gather_variables() %>%
head(10)## # A tibble: 10 × 5
## # Groups: .variable [1]
## .chain .iteration .draw .variable .value
## <int> <int> <int> <chr> <dbl>
## 1 1 1 1 overall_mean 0.0672
## 2 1 2 2 overall_mean 0.0361
## 3 1 3 3 overall_mean 1.17
## 4 1 4 4 overall_mean 0.378
## 5 1 5 5 overall_mean 0.359
## 6 1 6 6 overall_mean 0.332
## 7 1 7 7 overall_mean 0.226
## 8 1 8 8 overall_mean 0.107
## 9 1 9 9 overall_mean 0.225
## 10 1 10 10 overall_mean -0.0395But again, this approach does not handle variable indices for us
automatically, so using spread_draws() and
gather_draws() is generally recommended unless you do not
have variable indices to worry about.
Selecting variables using regular expressions
You can use regular expressions in the specifications passed to
spread_draws() and gather_draws() to match
multiple columns by passing regex = TRUE. Our example fit
contains variables named condition_mean[i] and
condition_zoffset[i]. We could extract both using a single
regular expression:
m %>%
spread_draws(`condition_.*`[condition], regex = TRUE) %>%
head(10)## # A tibble: 10 × 6
## # Groups: condition [1]
## condition condition_mean condition_zoffset .chain .iteration .draw
## <fct> <dbl> <dbl> <int> <int> <int>
## 1 A 0.00544 -0.0237 1 1 1
## 2 A -0.0836 -0.0468 1 2 2
## 3 A 0.0324 -0.827 1 3 3
## 4 A 0.113 -0.260 1 4 4
## 5 A 0.157 -0.207 1 5 5
## 6 A 0.218 -0.132 1 6 6
## 7 A 0.276 0.0642 1 7 7
## 8 A 0.0130 -0.0823 1 8 8
## 9 A 0.152 -0.0611 1 9 9
## 10 A 0.192 0.200 1 10 10This result is equivalent in this case to
spread_draws(c(condition_mean, condition_zoffset)[condition]),
but does not require us to list each variable explicitly—this can be
useful, for example, in models with naming schemes like
b_[some name] for coefficients.
Drawing fit curves with uncertainty
To demonstrate drawing fit curves with uncertainty, let’s fit a
slightly naive model to part of the mtcars dataset using
brms::brm():
m_mpg = brm(
mpg ~ hp * cyl,
data = mtcars,
file = "models/tidybayes_m_mpg.rds" # cache model (can be removed)
)We can draw fit curves with probability bands using
add_epred_draws() and
ggdist::stat_lineribbon():
mtcars %>%
group_by(cyl) %>%
data_grid(hp = seq_range(hp, n = 51)) %>%
add_epred_draws(m_mpg) %>%
ggplot(aes(x = hp, y = mpg, color = ordered(cyl))) +
stat_lineribbon(aes(y = .epred)) +
geom_point(data = mtcars) +
scale_fill_brewer(palette = "Greys") +
scale_color_brewer(palette = "Set2")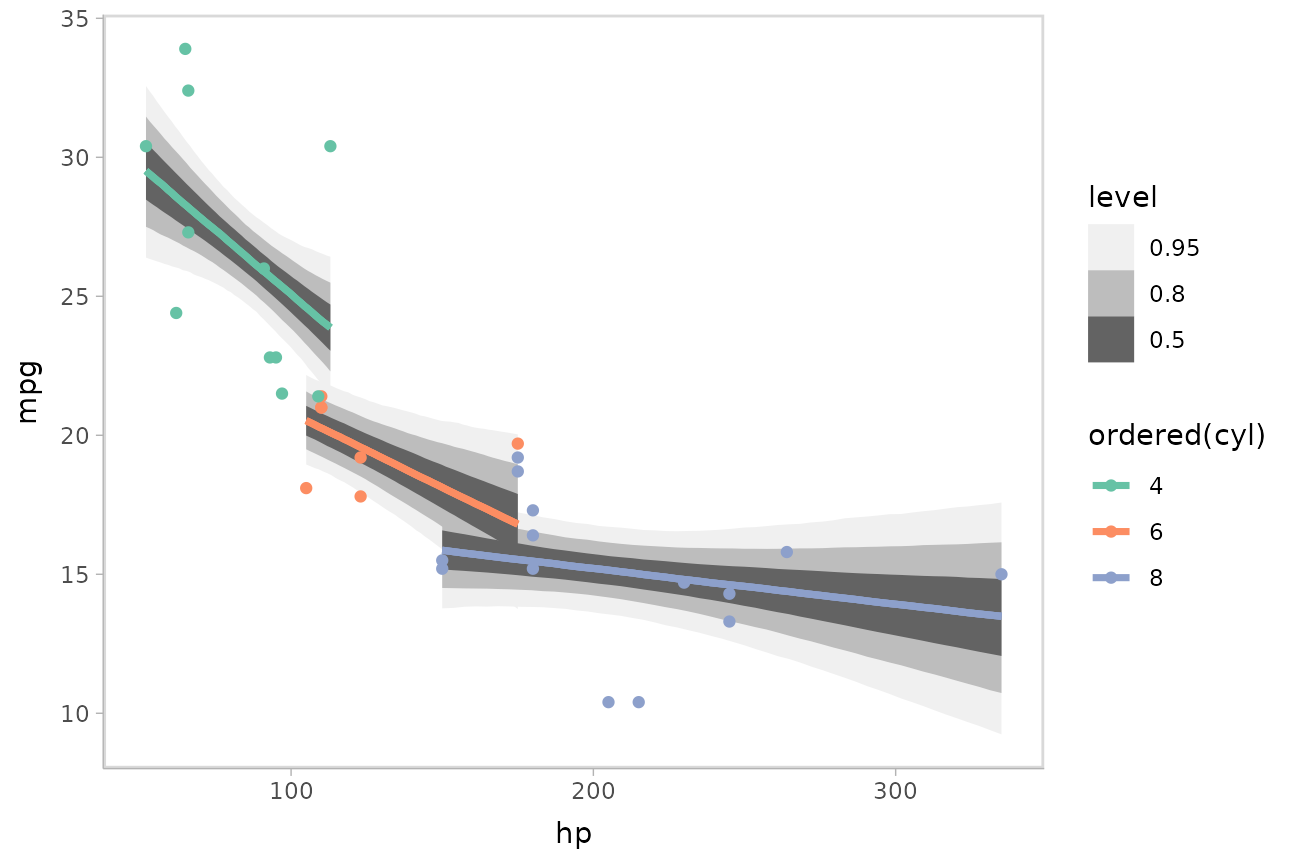
Or we can sample a reasonable number of fit lines (say 100) and overplot them:
mtcars %>%
group_by(cyl) %>%
data_grid(hp = seq_range(hp, n = 101)) %>%
# NOTE: this shows the use of ndraws to subsample within add_epred_draws()
# ONLY do this IF you are planning to make spaghetti plots, etc.
# NEVER subsample to a small sample to plot intervals, densities, etc.
add_epred_draws(m_mpg, ndraws = 100) %>%
ggplot(aes(x = hp, y = mpg, color = ordered(cyl))) +
geom_line(aes(y = .epred, group = paste(cyl, .draw)), alpha = .1) +
geom_point(data = mtcars) +
scale_color_brewer(palette = "Dark2")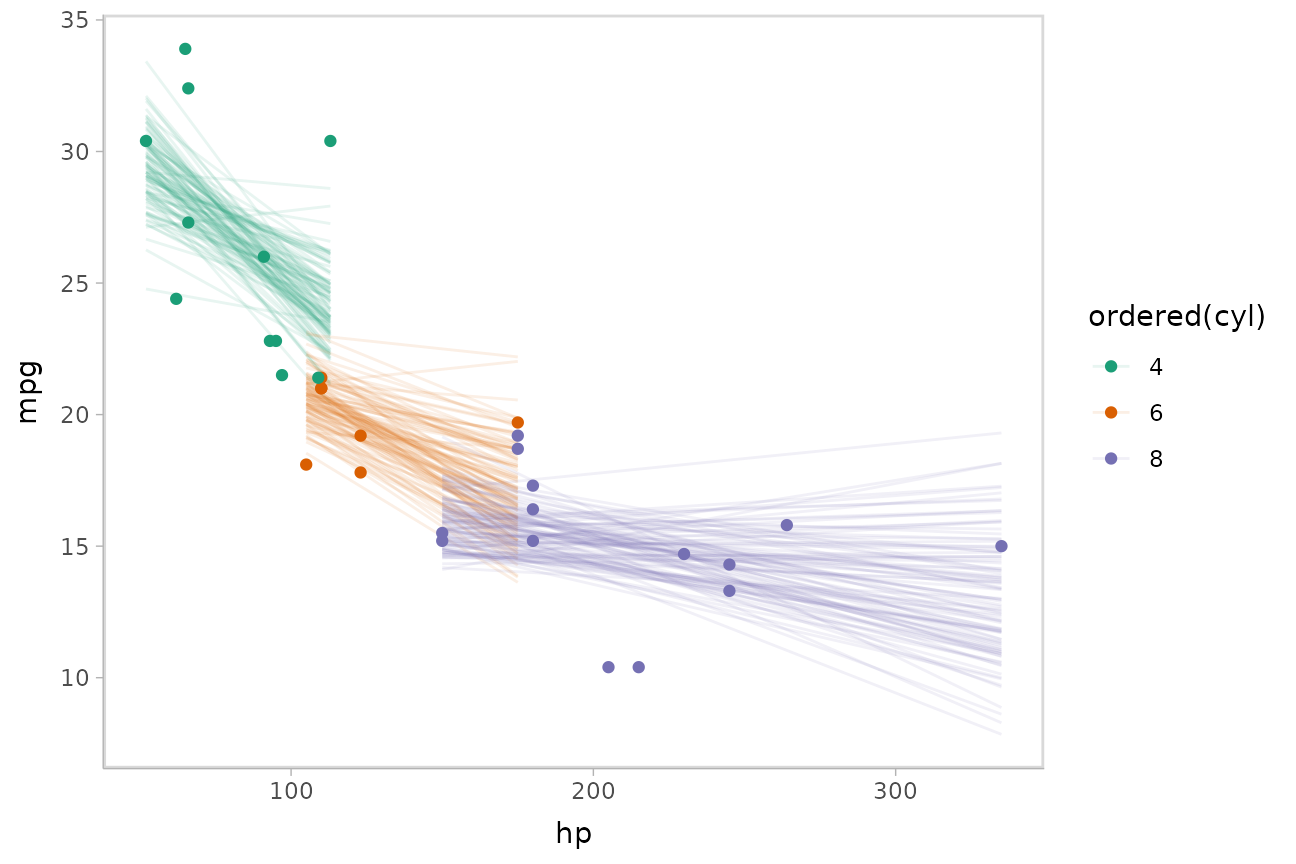
For more examples of fit line uncertainty, see the corresponding
sections in vignette("tidy-brms") or
vignette("tidy-rstanarm").
Compatibility with other packages
Compatibility of point_interval with
broom::tidy: A model comparison example
Combining to_broom_names() with median_qi()
(or more generally, the point_interval() family of
functions) makes it easy to compare results against models supported by
broom::tidy(). For example, let’s compare our model’s fits
for conditional means against an ordinary least squares (OLS)
regression:
m_linear = lm(response ~ condition, data = ABC)Combining emmeans::emmeans with
broom::tidy, we can generate tidy-format summaries of
conditional means from the above model:
linear_results = m_linear %>%
emmeans::emmeans(~ condition) %>%
tidy(conf.int = TRUE) %>%
mutate(model = "OLS")
linear_results## # A tibble: 5 × 9
## condition estimate std.error df conf.low conf.high statistic p.value model
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 A 0.182 0.173 45 -0.167 0.530 1.05 3.00e- 1 OLS
## 2 B 1.01 0.173 45 0.665 1.36 5.85 5.13e- 7 OLS
## 3 C 1.87 0.173 45 1.53 2.22 10.8 4.15e-14 OLS
## 4 D 1.03 0.173 45 0.678 1.38 5.93 3.97e- 7 OLS
## 5 E -0.935 0.173 45 -1.28 -0.586 -5.40 2.41e- 6 OLSWe can derive corresponding fits from our model:
bayes_results = m %>%
spread_draws(condition_mean[condition]) %>%
median_qi(estimate = condition_mean) %>%
to_broom_names() %>%
mutate(model = "Bayes")
bayes_results## # A tibble: 5 × 8
## condition estimate conf.low conf.high .width .point .interval model
## <fct> <dbl> <dbl> <dbl> <dbl> <chr> <chr> <chr>
## 1 A 0.199 -0.142 0.549 0.95 median qi Bayes
## 2 B 1.01 0.651 1.34 0.95 median qi Bayes
## 3 C 1.84 1.48 2.19 0.95 median qi Bayes
## 4 D 1.02 0.681 1.37 0.95 median qi Bayes
## 5 E -0.890 -1.23 -0.529 0.95 median qi BayesHere, to_broom_names() will convert .lower
and .upper into conf.low and
conf.high so the names of the columns we need to make the
comparison (condition, estimate,
conf.low, and conf.high) all line up easily.
This makes it simple to combine the two tidy data frames together using
bind_rows, and plot them:
bind_rows(linear_results, bayes_results) %>%
mutate(condition = fct_rev(condition)) %>%
ggplot(aes(y = condition, x = estimate, xmin = conf.low, xmax = conf.high, color = model)) +
geom_pointinterval(position = position_dodge(width = .3))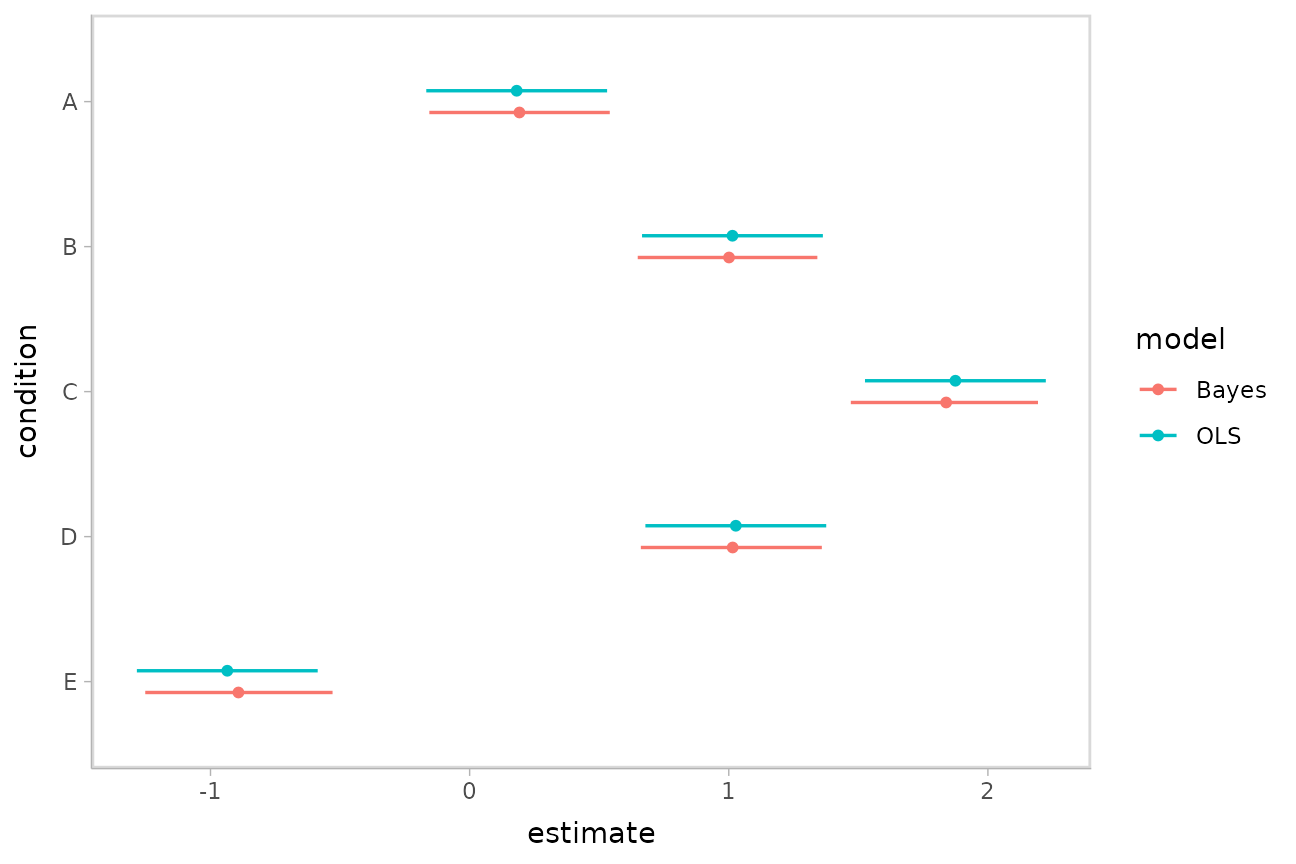
Observe the shrinkage towards the overall mean in the Bayesian model compared to the OLS model.
Compatibility with bayesplot using
unspread_draws and ungather_draws
Functions from other packages might expect draws in the form of a
data frame or matrix with variables as columns and draws as rows. That
is the format returned by tidy_draws(), but not by
gather_draws() or spread_draws(), which split
indices from variables out into columns.
It may be desirable to use the spread_draws() or
gather_draws() functions to transform your draws in some
way, and then convert them back into the draw
variable format to pass them into functions from other packages, like
bayesplot. The unspread_draws() and
ungather_draws() functions invert
spread_draws() and gather_draws() to return a
data frame with variable column names that include indices in them and
draws as rows.
As an example, let’s re-do the previous example of
compare_levels(), but use
bayesplot::mcmc_areas() to plot the results instead of
ggdist::stat_eye(). First, the result of
compare_levels() looks like this:
m %>%
spread_draws(condition_mean[condition]) %>%
compare_levels(condition_mean, by = condition) %>%
head(10)## # A tibble: 10 × 5
## # Groups: condition [1]
## .chain .iteration .draw condition condition_mean
## <int> <int> <int> <chr> <dbl>
## 1 1 1 1 B - A 1.02
## 2 1 2 2 B - A 0.957
## 3 1 3 3 B - A 0.921
## 4 1 4 4 B - A 0.881
## 5 1 5 5 B - A 0.676
## 6 1 6 6 B - A 0.599
## 7 1 7 7 B - A 0.813
## 8 1 8 8 B - A 0.940
## 9 1 9 9 B - A 0.750
## 10 1 10 10 B - A 0.794To get a version we can pass to bayesplot::mcmc_areas(),
all we need to do is invert the spread_draws() call we
started with:
m %>%
spread_draws(condition_mean[condition]) %>%
compare_levels(condition_mean, by = condition) %>%
unspread_draws(condition_mean[condition]) %>%
head(10)## # A tibble: 10 × 13
## .chain .iteration .draw `condition_mean[B - A]` `condition_mean[C - A]` condition_mean[C - B…¹ condition_mean[D - A…²
## <int> <int> <int> <dbl> <dbl> <dbl> <dbl>
## 1 1 1 1 1.02 1.84 0.814 1.25
## 2 1 2 2 0.957 1.86 0.906 1.20
## 3 1 3 3 0.921 1.76 0.838 1.13
## 4 1 4 4 0.881 1.96 1.08 0.925
## 5 1 5 5 0.676 1.95 1.27 0.904
## 6 1 6 6 0.599 1.75 1.15 0.693
## 7 1 7 7 0.813 1.14 0.331 0.754
## 8 1 8 8 0.940 1.61 0.673 1.01
## 9 1 9 9 0.750 1.78 1.03 0.968
## 10 1 10 10 0.794 1.72 0.927 0.834
## # ℹ abbreviated names: ¹`condition_mean[C - B]`, ²`condition_mean[D - A]`
## # ℹ 6 more variables: `condition_mean[D - B]` <dbl>, `condition_mean[D - C]` <dbl>, `condition_mean[E - A]` <dbl>,
## # `condition_mean[E - B]` <dbl>, `condition_mean[E - C]` <dbl>, `condition_mean[E - D]` <dbl>We can pass that into bayesplot::mcmc_areas() directly.
The drop_indices = TRUE argument to
unspread_draws() indicates that .chain,
.iteration, and .draw should not be included
in the output:
m %>%
spread_draws(condition_mean[condition]) %>%
compare_levels(condition_mean, by = condition) %>%
unspread_draws(condition_mean[condition], drop_indices = TRUE) %>%
bayesplot::mcmc_areas()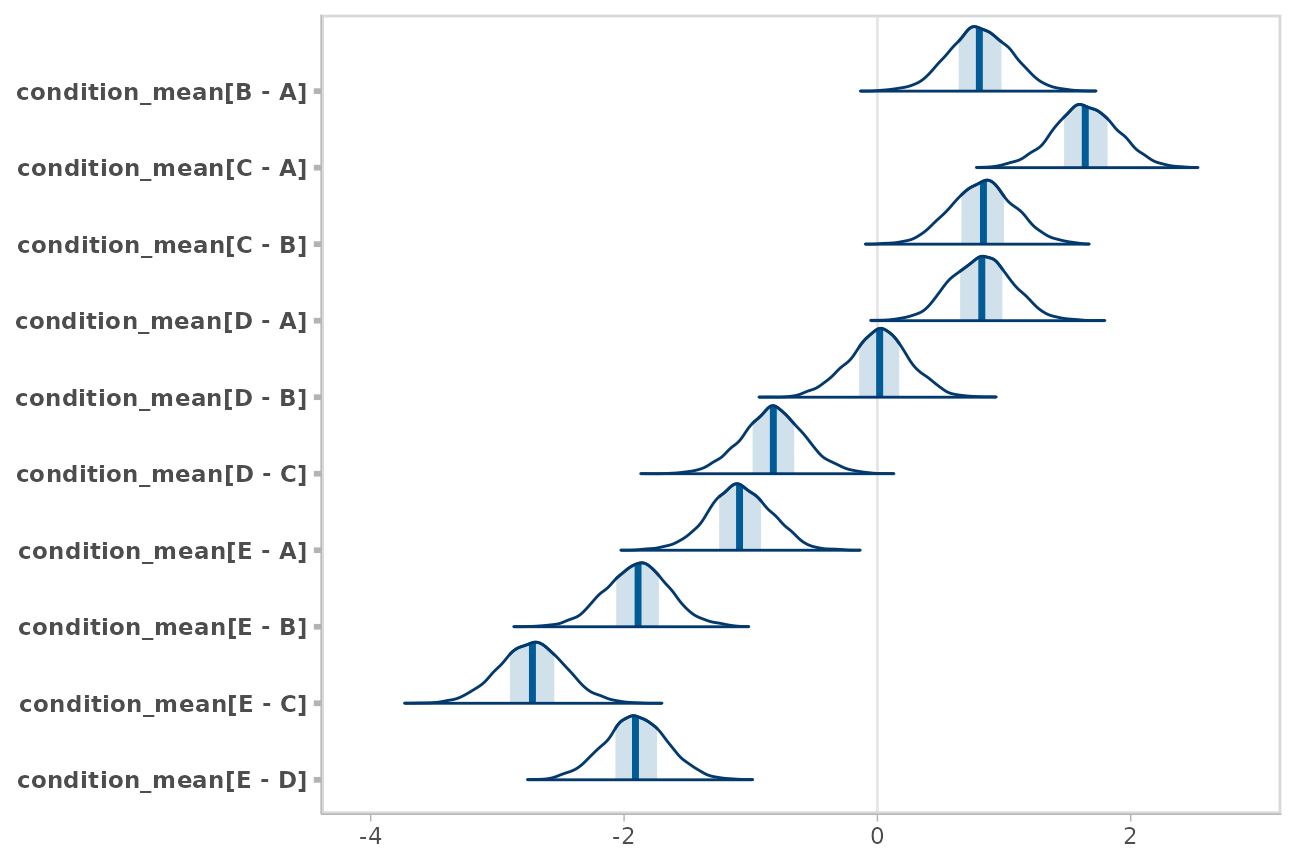
If you are instead working with tidy draws generated by
gather_draws() or gather_variables(), the
ungather_draws() function will transform those draws into
the draw
variable format. It has the same syntax as
unspread_draws().
Compatibility with emmeans (formerly
lsmeans)
The emmeans::emmeans() function provides a convenient
syntax for generating marginal estimates from a model, including
numerous types of contrasts. It also supports some Bayesian modeling
packages, like MCMCglmm, rstanarm, and
brms. However, it does not provide draws in a tidy format.
The gather_emmeans_draws() function converts output from
emmeans into a tidy format, keeping the
emmeans reference grid and adding a .value
column with long-format draws.
(Another approach to using emmeans contrast methods is
to use emmeans_comparison() to convert emmeans contrast
methods into comparison functions that can be used with
tidybayes::compare_levels(). See the documentation of
emmeans_comparison() for more information).
Using rstanarm or brms
Both rstanarm and brms behave similarly
when used with emmeans::emmeans(). The example below uses
rstanarm, but should work similarly for
brms.
Given this rstanarm model:
m_rst = stan_glm(response ~ condition, data = ABC)We can use emmeans::emmeans() to get conditional means
with uncertainty:
## # A tibble: 5 × 7
## condition .value .lower .upper .width .point .interval
## <fct> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 A 0.187 -0.158 0.526 0.95 median qi
## 2 B 1.01 0.659 1.36 0.95 median qi
## 3 C 1.87 1.52 2.22 0.95 median qi
## 4 D 1.03 0.683 1.39 0.95 median qi
## 5 E -0.937 -1.28 -0.592 0.95 median qiOr emmeans::emmeans() with
emmeans::contrast() to do all pairwise comparisons:
m_rst %>%
emmeans::emmeans( ~ condition) %>%
emmeans::contrast(method = "pairwise") %>%
gather_emmeans_draws() %>%
median_qi()## # A tibble: 10 × 7
## contrast .value .lower .upper .width .point .interval
## <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 A - B -0.822 -1.32 -0.324 0.95 median qi
## 2 A - C -1.68 -2.18 -1.21 0.95 median qi
## 3 A - D -0.837 -1.33 -0.347 0.95 median qi
## 4 A - E 1.13 0.615 1.61 0.95 median qi
## 5 B - C -0.863 -1.36 -0.339 0.95 median qi
## 6 B - D -0.0158 -0.521 0.499 0.95 median qi
## 7 B - E 1.95 1.45 2.45 0.95 median qi
## 8 C - D 0.843 0.342 1.34 0.95 median qi
## 9 C - E 2.81 2.33 3.31 0.95 median qi
## 10 D - E 1.97 1.46 2.46 0.95 median qiSee the documentation for emmeans::pairwise.emmc() for a
list of the numerous contrast types supported by
emmeans::emmeans().
As before, we can plot the results instead of using a table:
m_rst %>%
emmeans::emmeans( ~ condition) %>%
emmeans::contrast(method = "pairwise") %>%
gather_emmeans_draws() %>%
ggplot(aes(x = .value, y = contrast)) +
stat_halfeye()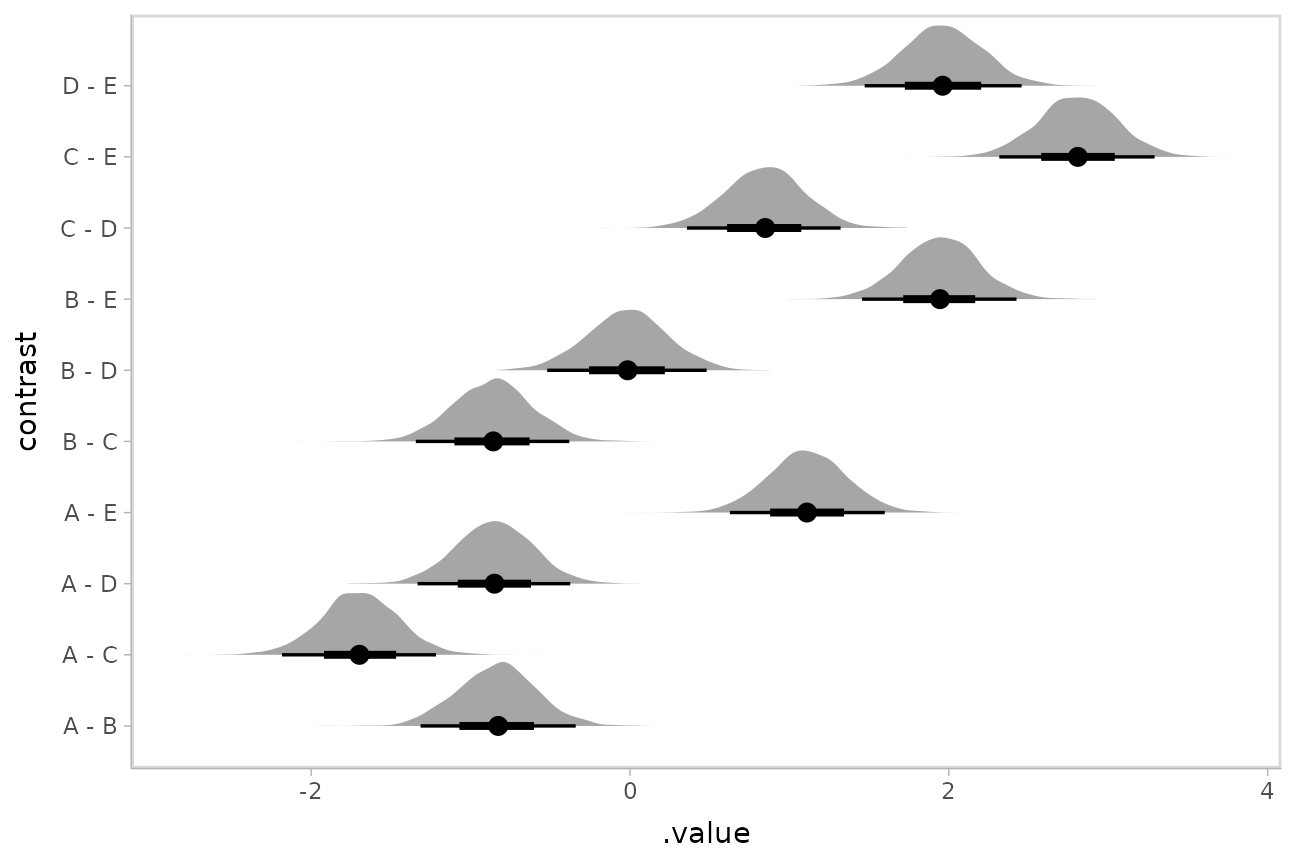
gather_emmeans_draws() also supports
emm_list objects, which contain estimates from different
reference grids (see emmeans::ref_grid() for more
information on reference grids). An additional column with the default
name of .grid is added to indicate the reference grid for
each row in the output:
## # A tibble: 15 × 9
## condition contrast .grid .value .lower .upper .width .point .interval
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 A NA emmeans 0.187 -0.158 0.526 0.95 median qi
## 2 B NA emmeans 1.01 0.659 1.36 0.95 median qi
## 3 C NA emmeans 1.87 1.52 2.22 0.95 median qi
## 4 D NA emmeans 1.03 0.683 1.39 0.95 median qi
## 5 E NA emmeans -0.937 -1.28 -0.592 0.95 median qi
## 6 NA A - B contrasts -0.822 -1.32 -0.324 0.95 median qi
## 7 NA A - C contrasts -1.68 -2.18 -1.21 0.95 median qi
## 8 NA A - D contrasts -0.837 -1.33 -0.347 0.95 median qi
## 9 NA A - E contrasts 1.13 0.615 1.61 0.95 median qi
## 10 NA B - C contrasts -0.863 -1.36 -0.339 0.95 median qi
## 11 NA B - D contrasts -0.0158 -0.521 0.499 0.95 median qi
## 12 NA B - E contrasts 1.95 1.45 2.45 0.95 median qi
## 13 NA C - D contrasts 0.843 0.342 1.34 0.95 median qi
## 14 NA C - E contrasts 2.81 2.33 3.31 0.95 median qi
## 15 NA D - E contrasts 1.97 1.46 2.46 0.95 median qiUsing MCMCglmm
Let’s do the same example as above again, this time using
MCMCglmm::MCMCglmm() instead of rstanarm. The
only different when using MCMCglmm() is that to use
MCMCglmm() with emmeans() you must also pass
the original data used to fit the model to the emmeans()
call (see vignette("models", package = "emmeans")) for more
information).
First, we’ll fit the model:
# MCMCglmm does not support tibbles directly,
# so we convert ABC to a data.frame on the way in
m_glmm = MCMCglmm::MCMCglmm(response ~ condition, data = as.data.frame(ABC))Now we can use emmeans() and
gather_emmeans_draws() exactly as we did with
rstanarm, but we need to include a data
argument in the emmeans() call:
m_glmm %>%
emmeans::emmeans( ~ condition, data = ABC) %>%
emmeans::contrast(method = "pairwise") %>%
gather_emmeans_draws() %>%
ggplot(aes(x = .value, y = contrast)) +
stat_halfeye()